Nikolai Slavov
Associate Professor, Bioengineering
Founding Director, Parallel Squared Technology Institute
Allen Distinguished Investigator, Allen Frontiers Group
Director, Single-Cell Proteomics Center
Affiliated Faculty, Biology
Affiliated Faculty, Chemistry and Chemical Biology
Office
- Mugar Life Sciences, Office 334
- 617.373.6405
Research Focus
Single-cell proteomics, immunology, cancer drug resistance, translational regulation, quantitative systems biology, mass-spectrometry, macrophage polarization, neurodegenerative diseases
About
Nikolai Slavov received undergraduate education from MIT and a doctoral degree from Princeton University for characterizing the coordination of cellular growth with gene expression and metabolism.
The Slavov laboratory pioneered experimental and computational methods for single-cell proteomics and used them to connect protein covariation across single cells to functional phenotypes, including macrophage polarization, emergence of drug resistance priming, early mammalian development, and stem cell differentiation. These technologies provided a foundation for establishing Parallel Squared Technology Institute (PTI).
Prof. Slavov organizes the annual single-cell proteomics conference and contributes to organizing other leading conferences, including NeurIPS.
Education
- PhD (2010), Botstein Laboratory, Princeton University
- BS (2004), Biology, Massachusetts Institute of Technology
Honors & Awards
- HUPO Discovery in Proteomic Sciences Award
- 2022 College of Engineering Faculty Fellow
- Allen Distinguished Investigator Award
- Excellence in Mentoring Award
- NIH Director’s New Innovator Award
- SPARC Award from the Broad Institute of MIT and Harvard
- Princeton University Dean’s Award
- IRCSET Postgraduate Research Fellowship
- Finalist in the Young European Entrepreneur Competition
- Princeton Graduate Fellowship
- MIT Undergraduate Fellowship
- Eureka Fellowship for Academic Excellence
- Bronze Medal in the 31st International Chemistry Olympiad
- National Diploma for Exceptional Achievements in Chemistry
Teaching Interests
Leadership Positions
- Founding director of Parallel Squared Technology Institute
- Established the annual Single-cell Proteomics conference
- Co-organized workshops at scientific conferences, including NeurIPS (Learning Meaningful Representations of Life), and HUPO
Professional Affiliations
- Founding director of Parallel Squared Technology Institute (PTI)
- Single-Cell Proteomics Center
- Barnett Institute of Chemical and Biological Analysis
- Broad Institute of MIT & Harvard
- American Society for Mass Spectrometry (ASMS)
- American Society for Cell Biology (ASCB)
- Genetics Society of America (GSA)
Research Overview
Single-cell proteomics, immunology, cancer drug resistance, translational regulation, quantitative systems biology, mass-spectrometry, macrophage polarization, neurodegenerative diseases
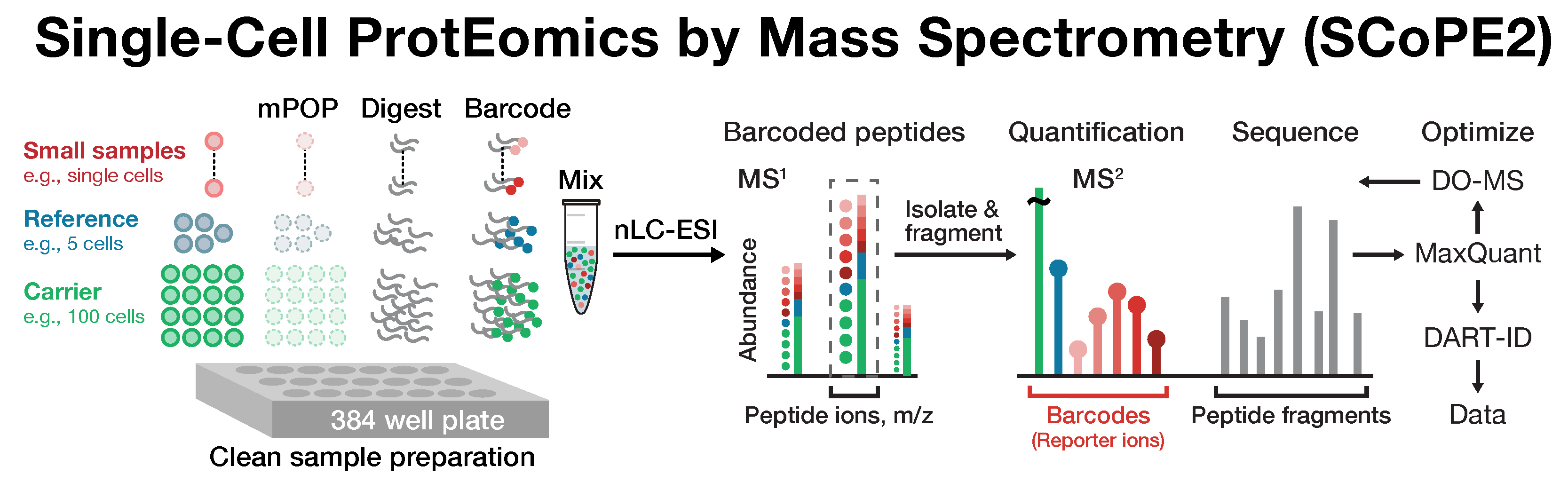
Single-cell proteomics by mass-spectrometry
Many biological processes stem from the coordinated interactions of molecularly and functionally diverse cells. However, this diversity is relatively unexplored at the proteome level because of the limitations of conventional affinity-based reagents for quantifying proteins in single cells. To alleviate these limitations, in 2017 our laboratory introduced Single Cell ProtEomics by Mass Spectrometry (SCoPE-MS). Since then, we developed more powerful and fully automated methods, including SCoPE2, pSCoPE, and plexDIA.
Taking advantage of ideas for advancing data acquisition and interpretation, we developed next generation methods that increase the sensitivity, data completeness and flexibility of single-cell protein analysis. These allow prioritization of thousands of proteins and highly parallel analysis of both single-cells and peptides. All of these methods can be implemented using accessible commercial equipment.
Ribosome-mediated translational regulation
All living cells must coordinate their metabolism, growth, division, and differentiation with their gene expression. Gene expression is regulated at multiple layers, from histone modifications (histone code) through RNA processing to protein degradation. While most layers are extensively studied, the regulatory role of specialized ribosomes (ribosome code) is largely unexplored. Such specialization has been suggested by the differential transcription of ribosomal proteins (RPs) and by the observation that mutations of RPs have highly specific phenotypes; particular RP mutations can cause diseases, known as ribosomopathies, and affect selectively the synthesis of some proteins but not of others. This selectivity and the differential RP transcription raise the hypothesis that cells may build specialized ribosomes with different stoichiometries among RPs as a means of regulating protein synthesis.
While the existence of specialized ribosomes has been hypothesized for decades, experimental and analytical roadblocks (such as the need for accurate quantification of homologous proteins and their modifications) have limited the evidence to only a few examples, e.g., the phosphorylation of RP S6. We developed methods to clear these roadblocks and obtained direct evidence for differential stoichiometry among core RPs in unperturbed yeast and mammalian stem cells and its fitness phenotypes. We aim to characterize ribosome specialization and its coordination with gene regulation, metabolism, and cell growth and differentiation. We want to understand quantitatively, conceptually, and mechanistically this coordination with emphasis on direct precision measurements of metabolic fluxes, protein synthesis and degradation rates in absolute units, molecules per cell per hour.
Slavov Laboratory
We aim to understand the rules governing emergent systems-level behavior and to use these rules to rationally engineer biological systems. We make quantitative measurements, often at the single-cell level, to test different conceptual frameworks and discriminate among different classes of models.
Selected Research Projects
Research Centers and Institutes
Department Research Areas
Selected Publications
Key Research Articles
- Huffman RG, Leduc A, Wichmann C, … and Slavov N.✉
- Leduc A, Huffman RG, Cantlon J, Khan S, Slavov N.✉
- Exploring functional protein covariation across single cells using nPOP Genome Biol 23, 261 (2022) preprint | PDF | nPOP Web | Data | Tutorials | Highlight by GEN | Perspective | Conference Talk
- Derks J, Leduc A, Wallmann G, Huffman G, Willetts M, Khan S, … Slavov N.✉
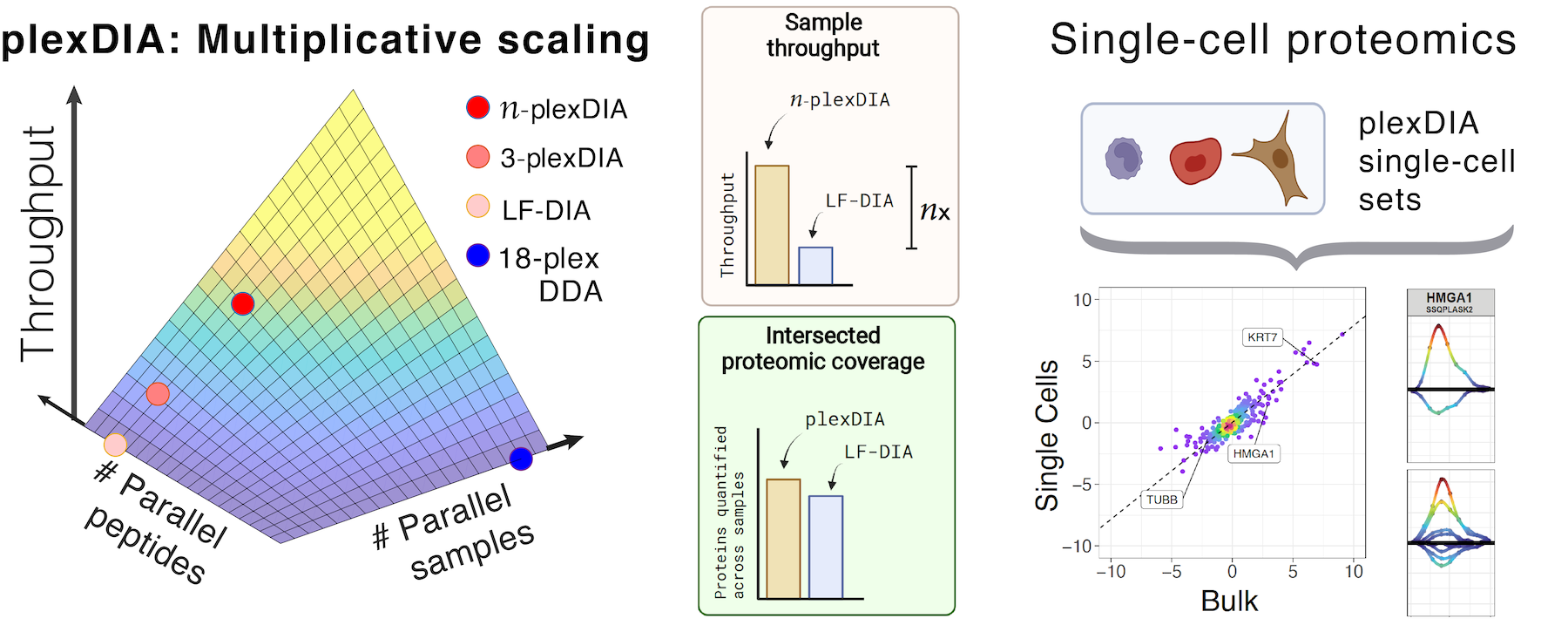
- Increasing the throughput of sensitive proteomics by plexDIA, Nature Biotechnology (2022) preprint | OA | PDF | plexDIA web | SCP2022 Talk | Nature Research Briefing | Nature Blog | Highlight by Nature Methods
- Budnik B, Levy E, Harmange G, Slavov N.✉
- SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation Genome Biology 19:161 (2018) preprint | PDF | Data | CSHL Talk | C&EN Highlight
Recent Research Articles
- Wallmann G., Leduc A., Slavov N.✉ (2023) Data-Driven Optimization of DIA Mass-Spectrometry by DO-MS J. Proteome Res. 22(10):3149-3158 OA | preprint | PDF | Code | Web
- Slavov N.✉ Single-cell proteomics: quantifying post-transcriptional regulation, Development 150, dev201492 (2023) PDF | Web Resources
- van den Berg P., Bérenger-Currias NM, Budnik B., Slavov N.✉, Semrau S.✉ (2023) Integration of a multi-omics stem cell differentiation dataset using a dynamical model PLoS Genet 19(5): e1010744 PDF | preprint | Data Web
- Huffman RG, Leduc A, Wichmann C, … and Slavov N.✉ (2023) Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics Nature Methods preprint | PDF | pSCoPE web | Data web | Talk | Nature Research Briefing (PDF)
- MacCoss MJ, Alfaro J, Faivre, D, Wu C, Wanunu M, Slavov N.✉ (2023) Sampling proteomes by emerging single-molecule and mass-spectrometry methods, Nature Methods 20, 339–346 preprint | PDF | SCP2022 Talk
- Gatto L, Aebersold R, Cox J, Demichev V, Derks J, …, Slavov N.✉ (2023) Initial recommendations for performing, benchmarking, and reporting single-cell proteomics experiments, Nature Methods preprint | PDF | Guidelines Web | Blog
- Derks J, Slavov N.✉ (2023) Strategies for increasing the depth and throughput of protein analysis by plexDIA, J. Proteome Res. preprint | PDF | Code | Web | YouTube Talk
- Leduc A, Huffman RG, Cantlon J, Khan S, Slavov N.✉ (2022) Exploring functional protein covariation across single cells using nPOP Genome Biol 23, 261 preprint | PDF | nPOP Web | Data | Tutorials | Highlight by GEN | Perspective | Conference Talk
- Derks J, Leduc A, Wallmann G, Huffman G, Willetts M, Khan S, Specht H, Ralser M, Demichev V, Slavov N.✉ (2022) Increasing the throughput of sensitive proteomics by plexDIA, Nature Biotechnology preprint | OA | PDF | plexDIA web | SCP2022 Talk | Nature Research Briefing | Nature Blog | Highlight by Nature Methods
- Slavov N.✉ (2022) Counting protein molecules for single-cell proteomics, Cell 185 (2) :232-234 PDF | OA
- Slavov N.✉ (2022) Learning from natural variation across the proteomes of single cells, PLoS Biology 20(1): e3001512 PDF | Website | Videos
- Slavov N.✉ (2021) Scaling up single-cell proteomics, Molecular & Cellular Proteomics PDF | Website | Recorded Workshop
- Petelski A, Emmott E, Leduc A, Huffman RG, Specht H, Perlman D, Slavov N.✉ (2021) Multiplexed single-cell proteomics using SCoPE2 Nature Protocols preprint | OA | PDF | Data web | Video Tutorials
- Slavov N.✉ (2021) Driving Single Cell Proteomics Forward with Innovation, J. of Proteome Res., doi: 10.1021/acs.jproteome.1c00639 | PDF
- Slavov N.✉ (2021) Increasing proteomics throughput, Nature Biotechnology PDF
- Slavov N.✉ et al. (2021) Voices of biotech research: Single-cell proteomics, Nature Biotechnology, 39, 281–286 PDF
- Specht H, Emmott E, Petelski A, Huffman RG, Perlman D, Serra M, Kharchenko P, Koller A. Slavov N.✉ (2021) Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2 Genome Biology preprint | PDF | Data | SCP2019 talk | SCoPE2 Web
- Specht H, Slavov N.✉ (2020) Optimizing accuracy and depth of protein quantification in experiments using isobaric carriers, J. of Proteome Res. Preprint | PDF | WebSite | SCP2020 talk | Code
- Petelski AA, Slavov N.✉ (2020) Analyzing ribosome remodeling in health and disease, Proteomics preprint PDF
- Slavov N.✉ (2020) Single-cell protein analysis by mass-spectrometry, Current Opinion in Chemical Biology, 60, 1-9 PDF
- Slavov N.✉ (2020) Unpicking the proteome in single cells, Science, 367 (6477) :512–513
- Slavov N.✉ et al. (2019) Voices in methods development: Single-cell proteomics Nature Methods, PDF
- Specht H, Emmott E, Koller T, Slavov N.✉ (2019) High-throughput single-cell proteomics quantifies the emergence of macrophage heterogeneity
bioRxiv DOI: 10.1101/665307 PDF | Data | SCP2019 talk | SCoPE2 Web - Chen A, Franks A, Slavov N.✉ (2019) DART-ID increases single-cell proteome coverage, PLoS Computational Biology, DOI: 10.1371/journal.pcbi.1007082 | PDF | RAW Data @ MassIVE | GitHub | DART-ID web
- Huffman RG, Specht H, Chen AT, Slavov N. (2019) DO-MS: Data-Driven Optimization of Mass Spectrometry Methods, J. of Proteome Res., DOI: 10.1021/acs.jproteome.9b00039 | DO-MS web | DO-MS @ GitHub
- Budnik B, Levy E, Harmange G, Slavov N.✉ (2018)
SCoPE-MS: mass-spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation
Genome Biology 19:161 preprint | PDF | Data | CSHL Talk | C&EN Highlight - Emmott EP, Jovanovic M, Slavov N.✉ (2018) Ribosome stoichiometry: from form to function, Trends in Biomedical Sciences, DOI: 10.1016/j.tibs.2018.10.009 PDF
- Malioutov D., Chen T., Jaffe J., Airoldi E., Budnik B., Slavov N. ✉ (2018) Quantifying homologous proteins and proteoforms, Molecular & Cellular Proteomics, DOI: 10.1101/168765 | PDF
- Specht H, Harmange G, Perlman DH, Emmott E, Niziolek Z, Budnik B, Slavov N.✉Automated sample preparation for high-throughput single-cell proteomics, bioRxiv, DOI: 10.1101/399774 PDF | RAW Data @ MassIVE | SCP2018 Talk
- Specht H. & Slavov N. (2018) Transformative opportunities for single cell proteomics J. of Proteome Res., 17 (8), 2565 – 2571
- Berg P., Budnik B., Slavov N., Semrau S. (2017) Dynamic post-transcriptional regulation during embryonic stem cell differentiation, bioRxiv, DOI: 10.1101/123497
- Franks A., Airoldi E.M., Slavov N. (2017) Post-transcriptional regulation across human tissues, PLoS Computational Biology, 13(5): e1005535, DOI: 10.1101/020206 | Highlight by The Scientist
- Slavov N. (2015) Making the most of peer review, eLife, 4:e12708
- Slavov N., Semrau S., Airoldi E., Budnik B., van Oudenaarden A., (2015) Differential Stoichiometry among Core Ribosomal Proteins, Cell Reports, 13(5), 865-873. (TiBS Spotlight)
- Malioutov D., Slavov N. (2014) Convex Total Least Squares, Journal of Machine Learning Research, W&CP, 32(1), 109-117.
- Slavov N., Budnik B., Schwab D., Airoldi E., van Oudenaarden A. (2014) Constant Growth Rate Can Be Supported by Decreasing Energy Flux and Increasing Aerobic Glycolysis, Cell Reports 7(3), 705-714.
- Slavov N., Carey, J., Linse, S. (2013) Calmodulin transduces Ca+2 oscillations into differential regulation of its target proteins, ACS Chemical Neuroscience, 4, 601-612.
- Slavov N., Botstein D. (2013) Decoupling Nutrient Signaling from Growth Rate Causes Aerobic Glycolysis and Deregulation of Cell Size and Gene Expression, Molecular Biology of the Cell, 24(2), 157-168,
- Slavov N., van Oudenaarden A. (2012) How to Regulate a Gene: To Repress or to Activate?, Molecular Cell, 46(5), 551-552.
- Slavov N., Airoldi E.M., van Oudenaarden A., Botstein D. (2012) A Conserved Cell Growth Cycle Can Account for the Environmental Stress Responses of Divergent Eukaryotes, Mol. Biol. Cell, vol. 23, no. 10
- Slavov N., Macinskas J., Caudy A., Botstein D. (2011) Metabolic Cycling without Cell Division Cycling in Respiring Yeast, PNAS, vol. 108, 19090-19095
- Slavov N., Botstein D. (2011) Coupling among Growth Rate Response, Metabolic Cycle and Cell Division Cycle in Yeast, Mol. Biol. Cell, vol. 22
- Slavov N. (2010) Inference of Sparse Networks with Unobserved Variables. Application to Gene Regulatory Networks, JMLR, W&CP vol. 9
- Slavov, N., Dawson, K. (2009) Correlation Signature of the Macroscopic States of the Gene Regulatory Network in Cancer, PNAS, vol. 106, no. 11 PDF
Feb 26, 2024
Northeastern Among Top 100 Universities for Utility Patents
Northeastern University, recognized for its excellence in innovation and entrepreneurship, has once again been named one of the top 100 universities worldwide for securing utility patents by the National Academy of Inventors.
Jan 22, 2024
Addressing Risk Factors and Potential Treatments of Alzheimer’s and Dementia
BioE Associate Professor Nikolai Slavov and Bouve Associate Professor Becky Briesacher provide insight into the differences between Alzheimer’s and dementia.
Dec 07, 2023
2023 Stanford University Annual Assessment of Author Citations
The following COE professors are among the top scientists worldwide selected by Stanford University representing the top 2% of the most-cited scientists with single-year impact in various disciplines. The selection is based on the top 100,000 by c-score (with and without self-citations) or a percentile rank of 2% or above. The list below includes those […]
Dec 04, 2023
Slowing the Progression of Alzheimer’s Disease
BioE Associate Professor Nikolai Slavov is investigating methods to slow the progression of Alzheimer’s, which is a specific form of dementia.
Oct 12, 2023
PhD Spotlight: Jason Derks, PhD’22, Bioengineering
Jason Derks’ PhD’22, bioengineering, research focused on developing new methods for high throughput proteomics and using them for quantifying the proteomes of single nuclei from macrophage cells responding to bacterial antigens. He developed technology he would call “plexDIA,” which he published in Nature Biotechnology in 2022.
Aug 10, 2023
New Methods of Analyzing Single Cell Proteomics
Allen Distinguished Investigator and BioE Associate Professor Nikolai Slavov was awarded a patent for “Mass spectrometry technique for single cell proteomics.”
Aug 08, 2023
Professor’s ‘Game-changing’ Alzheimer’s Research a Major Beneficiary of $50 Million Grant
A new research institute led by Allen Distinguished Investigator and BioE Associate Professor Nikolai Slavov is the principal beneficiary of a $50 million grant from two investors, Schmidt Futures and Citadel, to develop new techniques for studying Alzheimer’s disease.
Apr 04, 2023
Extending the Sensitivity, Consistency and Depth of Single-Cell Proteomics
The laboratory of Allen Distinguished Investigator and BioE Associate Professor Nikolai Slavov published their research on “Prioritized mass spectrometry increases the depth, sensitivity and data completeness of single-cell proteomics” in Nature Methods. Their method consistently analyzes thousands of prioritized peptides across all single cells (thus increasing data completeness) while maximizing instrument time spent analyzing identifiable peptides, thus increasing proteome depth.
Mar 30, 2023
Schmidt Futures Gives $50M for Life Science Research Including Single-Cell Proteomics Institute
The GenomeWeb article “Schmidt Futures Gives $50M for Life Science Research Including Single-Cell Proteomics Institute” highlights the Parallel Squared Technology Institute (PTI), a new non-profit research institute recently launched by Allen Distinguished Investigator and BioE Associate Professor Nikolai Slavov and former bioengineering students Aleksandra Petelski, PhD’22, and Harrison Specht, PhD’22.
Mar 17, 2023
PTI Launches to Scale Protein Analysis Towards Scientific and Medical Breakthroughs
Allen Distinguished Investigator and BioE Associate Professor Nikolai Slavov launched his new Parallel Squared Technology Institute (PTI) to catalyze a leap in biomedical research by enabling protein analysis with single-cell resolution at unprecedented throughput.